Our Primary Research Initiatives in Biosystems Innovations
Spotlight On The Disciplines Driving Sustainable Advancement
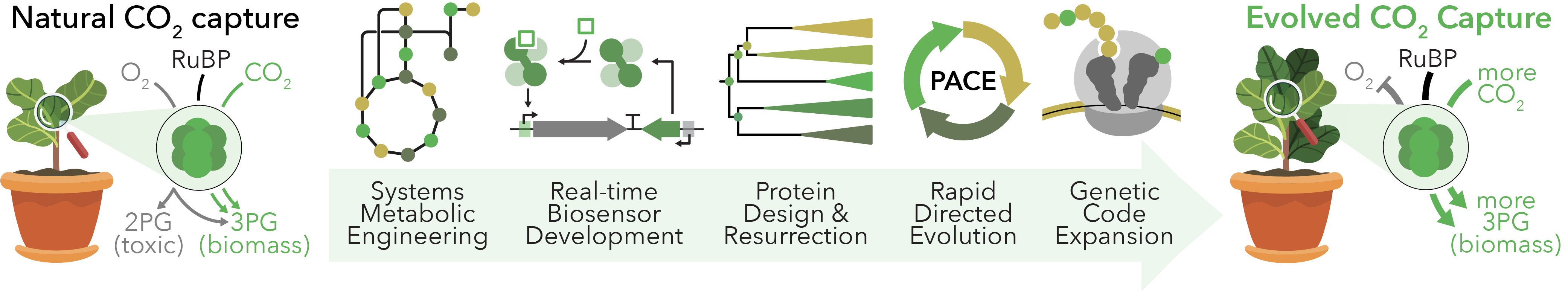
Carbon Capture
Climate change is the greatest threat affecting our planet. It has increased global temperatures, incentivized antibiotic resistance, intensified rain cycles, and will soon impact coastal communities through rising sea levels. This outlook is a consequence of centuries of atmospheric carbon dioxide (CO2) buildup at an alarming rate, which has prompted investigations into sustainable technologies for global CO2 management. Photosynthetic organisms may provide an immediate and sustainable means to affect this crisis through the enzyme RuBisCO, which captures atmospheric CO2 and fixes it into useful biomolecules. However, RuBisCO is an inefficient enzyme that cannot easily distinguish between CO2 and O2. We are combining strategies from in vivo metabolic engineering, in silico protein design, and high-throughput directed evolution to fundamentally reexamine biological CO2 capture and develop next-generation enzymes that effectively capture atmospheric CO2 for long-term climate correction strategies. Looking forward, our carbon-negative biosynthetic pathways may be used to directed convert airborne CO2 into fuels, foods, and materials.
Relevant Publications
Costello A, Badran AH.
Trends in Biotechnology 2020 PDF
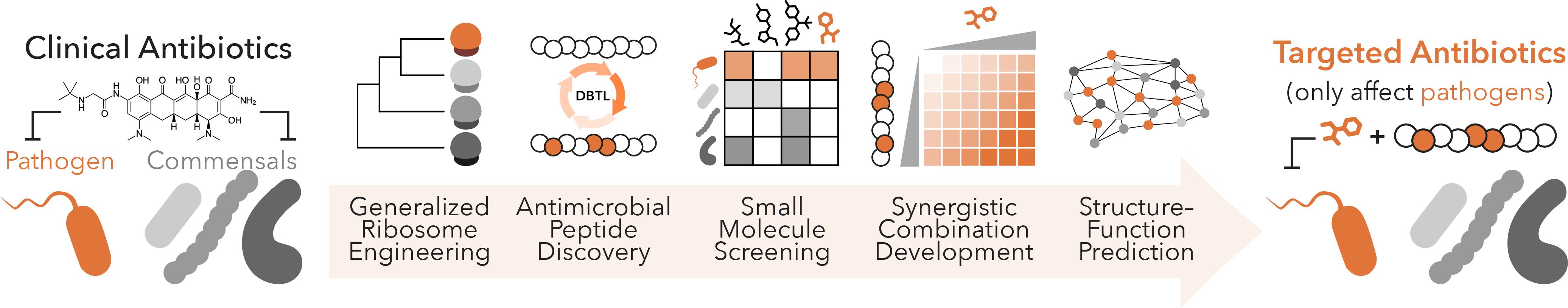
Targeted Antibiotics
Despite their medical utility, widespread antibiotic use has fueled the emergence of antimicrobial resistance (AMR). Catalyzed by the increasing global temperatures, pathogenic bacteria are adapting to frontline antibiotics more effectively than ever before, leading to higher AMR rates and worse patient prognosis. The current trajectory has made AMR the most severe threat to modern medicine and human health, where models predict that infectious diseases will become increasingly to treat worldwide. In ongoing work to address this, we are developing platforms that nominate next-generation antibiotics at an accelerated pace to effectively address AMR in human pathogens while mitigating damage to beneficial gut bacteria, thereby eliminating longstanding issues in addressing bystander bacterial depletion. We capitalize on new strategies to engineer translation machinery in vivo, large libraries of genetically encoded or synthetic small molecule libraries, and automated evolution methods to identify putative resistance mechanisms. These efforts will serve as a foundation for novel antibiotics discovery and identifying conditions that modulate AMR development.
Relevant Publications
Natural and Engineered Precision Antibiotics in the Context of Resistance
Johnston, CW, Badran AH.
Current Opinion in Chemical Biology 2022 — PDF
Directed Evolution of rRNA Improves Translation Kinetics and Recombinant Protein Yield
Liu F*, Bratulic S*, Costello AM*, Miettinen T, Badran AH.
Nature Communications 2021)
Kolber N, Fattal R, Bratulic S, Carver GD, Badran AH.
Nature Communications 2021 PDF / SI
Highlighted at the Broad Institute, Phys.org, and Chemical and Engineering News.
Johnston C, Badran AH, Collins JJ.
Nature Communications 2020 PDF / SI
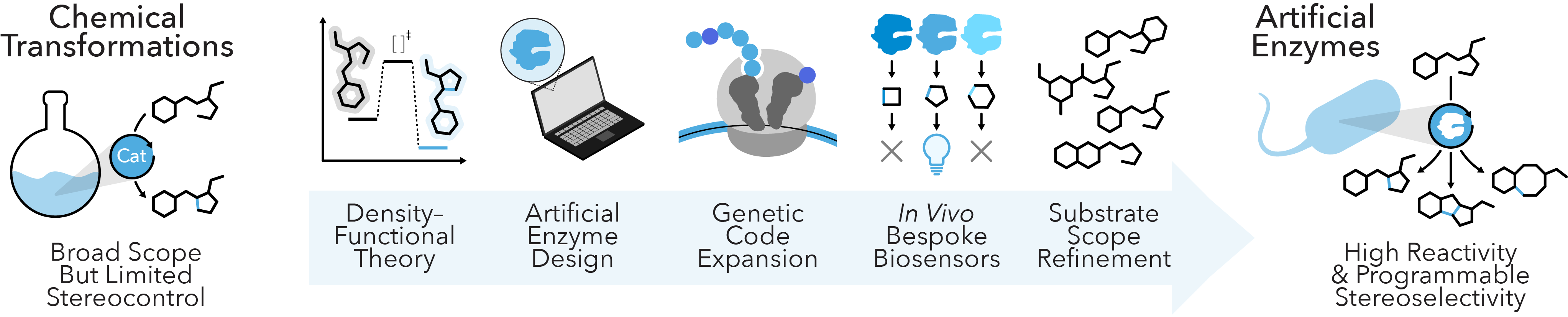
Artificial Enzymes
Enzymes enable exquisite chemistry yet can be limited in their transformational scope. Small molecule catalysts, on the other hand, can incorporate elements beyond the native building blocks of the cell to access new reactivities, but rarely afford the rate enhancements or enantioselectivities seen with enzymes. Seminal efforts to bridge this gap have relied on proteins with engineered cofactors to catalyze novel chemical transformations, but they remain restricted to a small subset of possible synthetic reactions. We’re overcoming these limitations by engineering new-to-nature enzymes that integrate synthetic ligands, which expand their chemical capabilities and yield efficient biological machinery for defined catalytic outcomes. We achieve this by integrating de novo enzyme design and bespoke in vivo biosensors with enhanced strategies for genetic code expansion, which together can uncover tailored biosynthetic enzymes with reactivities beyond those seen in nature. Our work will establish a new paradigm for artificial enzyme activities that catalyze researcher-defined reactions with programmable enantioselectivity.
Relevant Publications
DeBenedictis E*, Carver GD*, Chung C, Söll D, Badran AH.
Nature Communications 2021 PDF / SI
Highlighted at The Scripps Research Institute and Phys.Org.
Directed Evolution of rRNA Improves Translation Kinetics and Recombinant Protein Yield
Liu F*, Bratulic S*, Costello AM*, Miettinen T, Badran AH.
Nature Communications 2021)
Therapeutics Discovery
While human genetics has greatly furthered our understanding of disease states, conventional drug development processes continue to bottleneck the translation of these insights into urgently needed therapeutics. Rapidly translating these basic insights into therapeutics requires methods to quickly validate putative targets in disease to nominate potential liabilities, and uncover new molecules that engage and disrupt target function. To bridge the gap between target nomination and therapeutics development, we are developing a novel approach for molecular glue discovery, an exciting strategy to address key therapeutic targets in human cells. In nature, molecular glues are small molecules that have been refined through natural selection to modulate protein–protein interactions. Inspired by Nature, our lab is uncovering genetically-encodable de novo molecular glues that mediate new protein-protein interactions. We achieve this through genetic code expansion strategies to incorporate privileged protein-binding chemotypes alongside automated in vivo directed evolution methods to refine the structures, binding affinities, and specificities of new molecular glues.
Relevant Publications
Coming soon…


